This article is more than 1 year old
How a HPC array helps humanity destroy the Ebola virus
Big Iron makes everyone's lives better
Comment Deadly diseases like Ebola are frightening. They kill people in horrible ways and destroy the host communities’ ability to fight the spread of the virus. Agencies fighting the outbreak have had to understand where the outbreak is spreading, where communities in its path are most vulnerable and where to focus their treatment and containment efforts to ring-fence the affected areas.
Ebola virus
With such a fast-moving virus it’s been necessary to use computer simulations to model how the virus infects populations and assess how effective different actions, such as treatment station location, isolation measures, contact levels, hospital upgrades and hygiene education programs can be.
In other words, to assess where's best to concentrate resources in order to limit the numbers of people who will die from the Ebola epidemic.
What’s done is to use IT to simulate a human population in areas such as Liberia, Mali, Guinea and elsewhere, along with characteristics that encourage – and inhibit – Ebola’s spread. The aim is to create a disease spread model that is as close as possible to the actual West African outbreak’s history.
You redefine the model until it’s as close to what has happened as possible. Then you add in disease counter-measures, such as public hygiene education, travel restrictions and so forth, and predict what will then happen as a result of applying them.
This work has been carried out by the Virginia Bioinformatics Institute (VBI) with its 250 scientists and staff, who develop develop genomic, proteomic and bioinformatics tools, databases and computational models. They started work in July 2014 with the aim of developing a better understanding of how to respond to the Ebola outbreak which, as of February 2015, has infected 22,000 people and caused some 9,000 deaths.
The VBI staff worked with other agencies, such as the US Department of Defense’s Defense Threat Reduction Agency (DTRA), the American National Institutes of Health (NIH), the World Health Organization (WHO) and West Africa’s Ministries of Health (MOH) to forecast the disease’s spread and reduce and counter the risks of it moving outside infected regions into other parts of the world.
VBI scientists created an adaptable set of global synthetic populations with detailed demographics, family structures, travel patterns and activities to help model what could happen as the outbreak spreads. These synthetic populations mirror actual census, social, transit and telecommunications data patterns from the target populations.
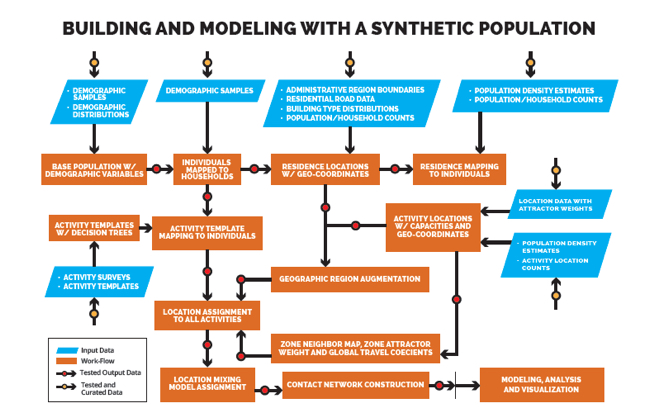
It’s complex IT, with millions of simulations applied to dozens of computational models and simulation engines to determine best how to respond on the ground. Some models require a lot of data while others have a constant stream of information requiring parallel processing. The faster this modelling can be done and its results communicated to field personnel, the more effective outbreak counter-measures will be.
Keith Bisset, a senior research scientist for VBI, said; “We built virtual cities on local, regional and global levels, infused with a tremendous amount of demographics collected from census, power, cell phone, transportation, social networks and other data. Our ability to expand our population framework enabled us to move to Ebola modelling very quickly.”
One model involved a synthetic population of the USA with events within it tracked over a 120-day model period (not real time). Some if the IT seems simply fantastic: creating this synthetic US population took 12 seconds and expanding it to a global scale would take all of six minutes. Then you run the simulations and that takes time, with the storage taking a beating. The faster the storage system can react, the better.
Bisset said: “Just one instance of our global population requires 10TB, and we might run [a] simulation 15 different times with variant conditions.”
The synthetic population was built up to number more than 750 million people. VBI uses its 2,500 core Shadowfax compute cluster to run the modelling work along with almost 1PB of storage. This was implemented as a cluster of two DDN GRIDScaler appliances using GPFS for parallel filesystem access.
The researchers had to scale up models quickly as events took place as when, for example the case count in Sierra Leone jumped exponentially in September 2014. Census data in parts of West Africa was lacking and so VBI structured models with social triggers and hooks based on behaviour. There was constant feedback which helped generate a massive amount of data.
Response time can be crucial. One example was when VBI received a call from the DOD on a Friday requesting input on where to place new emergency treatment units by Monday morning, when the military transport aircraft were taking off. Scientists looked at many variables over the weekend, including the road infrastructure in West Africa, and identified “hot spots” where additional outbreaks were thought to be imminent.
It’s satisfying to realise that our fairly dry storage industry and its goings-on can contribute to real-world emergency events like the Ebola outbreak. Ebola is now coming under control and having its malign effects reduced. Spinning platters can make a difference. ®
